Team Members:
- Saugat Tiwari
- Manish Bhattarai
- Suraj Raj Bohara
Project Supervisor:
Er. Kumar Prasun
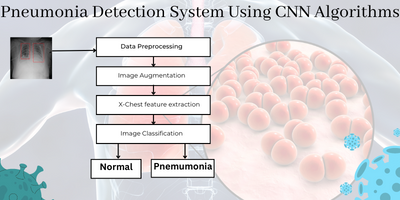
Pneumonia Detection System
Note from the editors: Towards Data Science is a Medium publication primarily based on the study of data science and machine learning. We are not health professionals or epidemiologists, and the opinions of this article should not be interpreted as professional advice. To learn more about the coronavirus pandemic, you can click here.
Pneumonia is a disease that affects the lungs and can cause symptoms such as coughing, chest pain, fever, sweating, fatigue, and loss of appetite. Machine learning algorithms can be used to detect the disease from images of Chest X-rays. The article discusses the use of InceptionV3 in detecting Pneumonia in Chest X-rays, and the entire code for training and testing the models as well as running the Flask app is available on my GitHub private repo.
Please get in touch with me directly on LinkedIn if you need the source code; I’ll include the link of my linkedin at the end of the blog.
The most common symptoms of pneumonia include coughing, chest pain, fever, sweating, fatigue, and loss of appetite. However, it is important to note that these symptoms can also be caused by other illnesses, and only a medical professional can accurately diagnose pneumonia. Other symptoms that people may experience include aches, pains, or difficulty in breathing. Most of these symptoms show signs of respiratory infections and lung abnormalities which can be detected by radiologists.
Thus, it is possible to use Machine Learning algorithms to detect the disease from images of Chest X-rays. Automated applications can be created to help support radiologists. This article is an attempt to use InceptionV3.
The entire code for training and testing the models as well as running the Flask app is available on my Github repository which is private now.
The Dataset
The dataset for the project was gathered from kaggle datasets:
Chest X-rayimages (5,863 images, in 2 categories) were obtained from: https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia
Algorithms used: InceptionV3 was trained on Chest X-rays. 90% of the images were used for training the models and the remaining 10% for testing the accuracy of the models.
Building the Model
The following code builds the InceptionV3 model for the Pneumonia Detector Flask App based on Chest X-rays. It initializes the weights for the model from the imagenet dataset and includes the top layer. It takes the input of shape (224, 224, 3) and flattens the output of the convolutional layers. It also applies a 50% dropout rate to the outputs and adds a dense output layer with softmax activation for the two classes (pneumonia or normal). Then, it sets the trainable attribute of the previous layers to False, compiles the model with categorical crossentropy as the loss function and the Adam optimizer, and applies data augmentation to the training set.
inception = InceptionV3(weights="imagenet", include_top=False,input_tensor=Input(shape=(224, 224, 3)))
outputs = inception.output
outputs = Flatten(name="flatten")(outputs)
outputs = Dropout(0.5)(outputs)
outputs = Dense(2, activation="softmax")(outputs)
model = Model(inputs=inception.input, outputs=outputs)
for layer in inception.layers:
layer.trainable = False
model.compile(
loss='categorical_crossentropy',
optimizer='adam',
metrics=['accuracy']
)
train_aug = ImageDataGenerator(
rotation_range=20,
width_shift_range=0.2,
height_shift_range=0.2,
horizontal_flip=True
)
We then added a Dropout layer to avoid overfitting and a Flatten layer to flatten all of our features. Finally, using the softmax function as the activation function, We added the Dense output layer. The trainable attribute of the preceding layers was set to False because the first half of the model has already been pretrained. By using the Adam optimizer and categorical crossentropy as the loss function, We finally assembled the model.
Training the Model
Training of the model was performed, with all the required parameters specified as follows:
history = model.fit(train_aug.flow(X_train, y_train, batch_size=32),
validation_data=(X_test, y_test),
validation_steps=len(X_test) / 32,
steps_per_epoch=len(X_train) / 32,
epochs=500)
As you can see, We have trained the model for 500 epochs with a batch size of 32 images.
Model Predictions
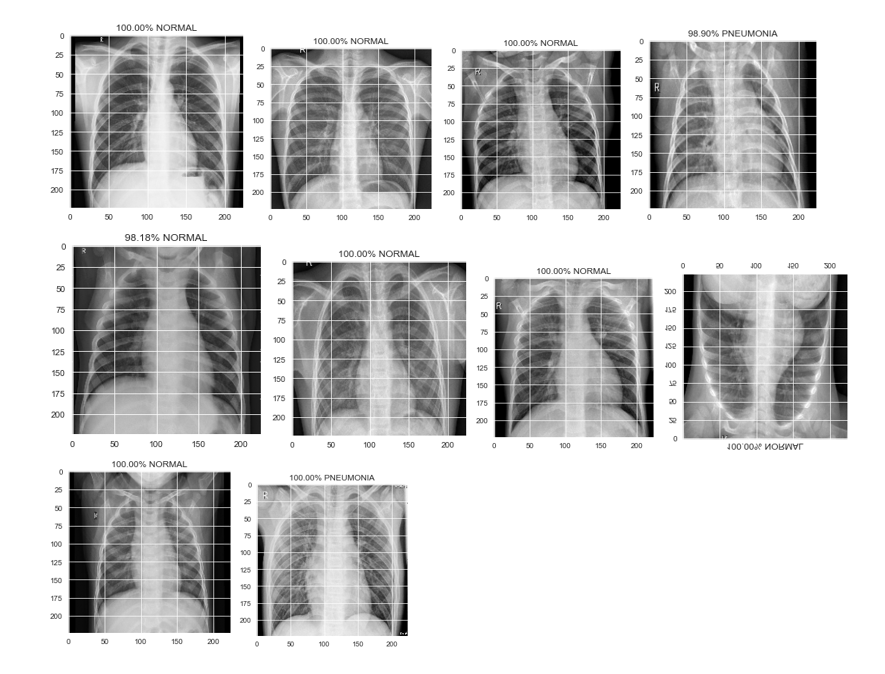
Predictions were generated by running the trained models on images of the test set. The predictions for the first 10 images of the dataset were plotted as shown below:
prediction=y_pred[0:10]
for index, probability in enumerate(prediction):
if probability[1] > 0.5:
plt.title('%.2f' % (probability[1]*100) + '% PNEUMONIA')
else:
plt.title('%.2f' % ((1-probability[1])*100) + '% NORMAL')
plt.imshow(X_test[index])
plt.show()
The following snippet shows the plots of first 10 predictions of Chest X-rays:
Evaluation & Results of Machine Learning
Following are a few important results and plots that help estimate the accuracy of the models and get insights into their performance.
Sample output of test set images
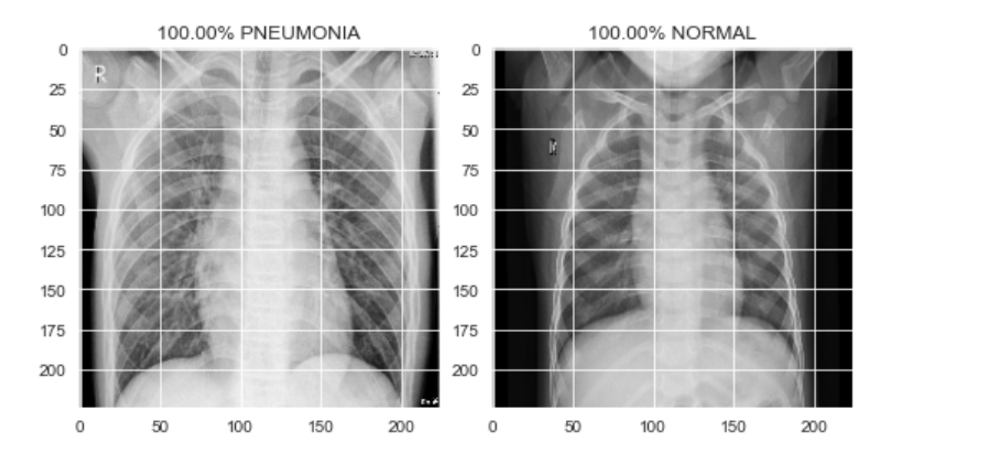
Fig: Sample output of test set images
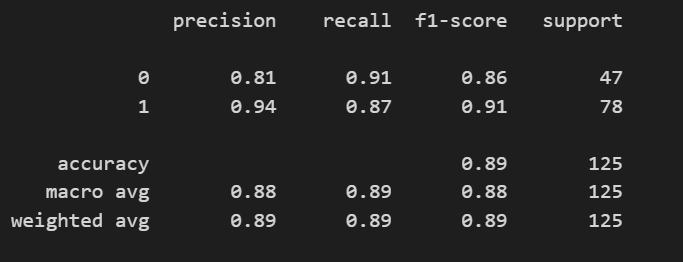
Classification Report
Classification Reports of InceptionV3 model for Chest X-rays
Confusion Matrix
Confusion Matrix of InceptionV3 model for Chest X-rays
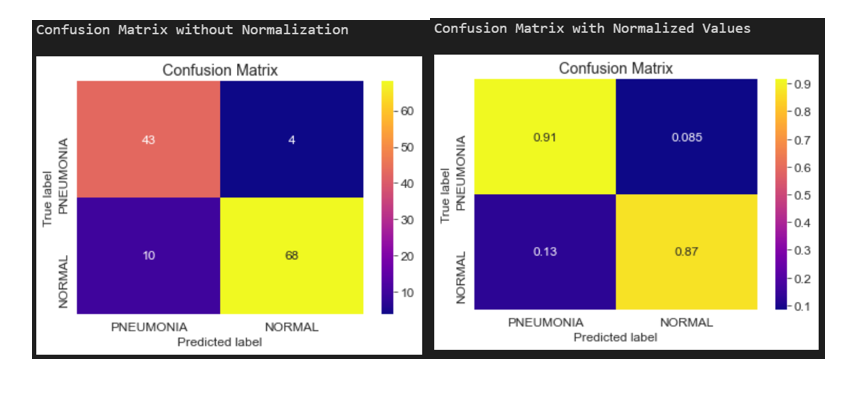
Fig: Confusion Matrix of InceptionV3 model for Chest X-rays
Building the Flask App
Now, We used the code I’d written thus far to create a Flask app by pasting specific segments of it into Flask functions. For example, the code for using the models to predict Chest X-rays looks like this:
def uploaded_chest():
if request.method == 'POST':
# check if the post request has the file part
if 'file' not in request.files:
flash('No file part')
return redirect(request.url)
file = request.files['file']
# if user does not select file, browser also
# submit a empty part without filename
if file.filename == '':
flash('No selected file')
return redirect(request.url)
if file:
# filename = secure_filename(file.filename)
file.save(os.path.join(app.config['UPLOAD_FOLDER'], 'upload_chest.jpg'))
inception_chest = load_model('models/inceptionv3_chest.h5')
image = cv2.imread('./flask app/assets/images/upload_chest.jpg') # read file
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB) # arrange format as per keras
image = cv2.resize(image,(150,150))
image = np.array(image) / 150
image = np.expand_dims(image, axis=1)
inception_pred = inception_chest.predict(image)
probability = inception_pred[0]
print("Predictions using InceptionV3 Algorithms:")
if probability[0] > 0.5:
inception_chest_pred = str('%.2f' % (probability[0]*100) + '% PNEUMONIA')
else:
inception_chest_pred = str('%.2f' % ((1-probability[0])*100) + '% NORMAL')
print(inception_chest_pred)
if __name__ == '__main__':
app.debug = True
app.run()
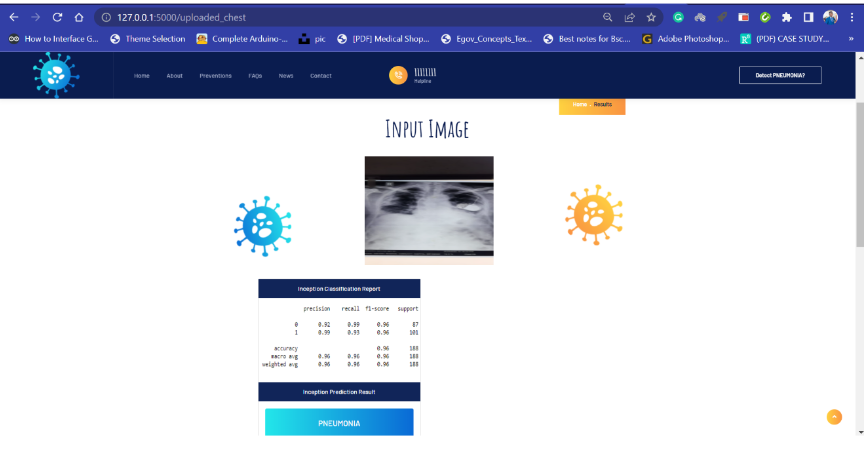
Screenshots of Flask App
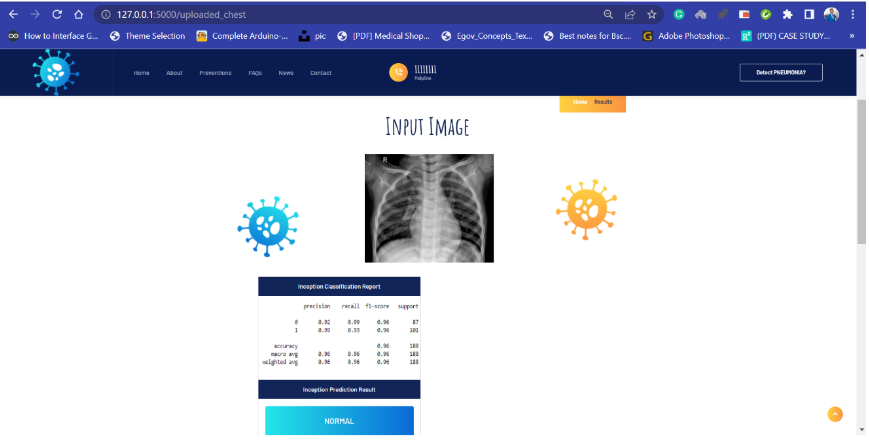
Source Code
The source code for the entire project, as well as the datasets, models, and flask app, is available on: GitHub private repo.
Conclusion
In conclusion, We would like to throw light on the fact that the analysis has been done on a limited dataset and that the results are preliminary. Medical validations have not been done on the approach and hence the results might differ from those observed in practical use cases.
In the future, We plan to improve the performance of the models by training them on more images and possibly including other factors like age, nationality, gender, etc. Furthermore, We encourage the readers of this article to experiment with the code on their own to increase the precision of the model.
We are generously obligated to our
supervisorEr. Kumar Prasun for his consistent help and direction all through this project. It was his significant recommendation and suggestion that helped us to adapt up to the rising impediments amid the improvement of this project.
Feel free to share this article with others if you found it useful. Thank you so much for reading.
Note: We do not have a medical background, and this project was created for a final-year college project and to demonstrate a concept.
Feel free to connect with me on other platforms:
GitHub: https://github.com/saugat86
LinkedIn: https://www.linkedin.com/in/saugattiwari/
Medium: saugattiwari.medium.com
Portfolio: https://saugaattiwari.com.np/